GENAVi offers rapid DEA using DESeq2 and gene set or pathway enrichment... | Download Scientific Diagram
SARTools: A DESeq2- and EdgeR-Based R Pipeline for Comprehensive Differential Analysis of RNA-Seq Data | PLOS ONE
![PDF] SARTools : a DESeq 2-and edgeR-based R pipeline for comprehensive differential analysis of RNA-Seq data | Semantic Scholar PDF] SARTools : a DESeq 2-and edgeR-based R pipeline for comprehensive differential analysis of RNA-Seq data | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/34c4616e3d7d5fd4adc257bdaced7178f4a40883/6-Figure2-1.png)
PDF] SARTools : a DESeq 2-and edgeR-based R pipeline for comprehensive differential analysis of RNA-Seq data | Semantic Scholar
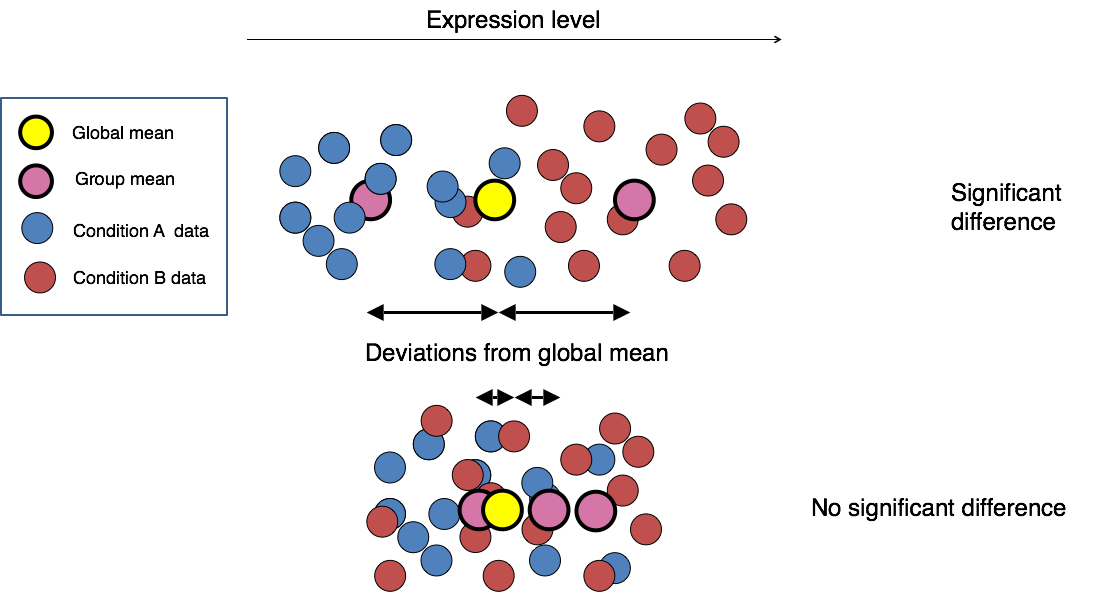
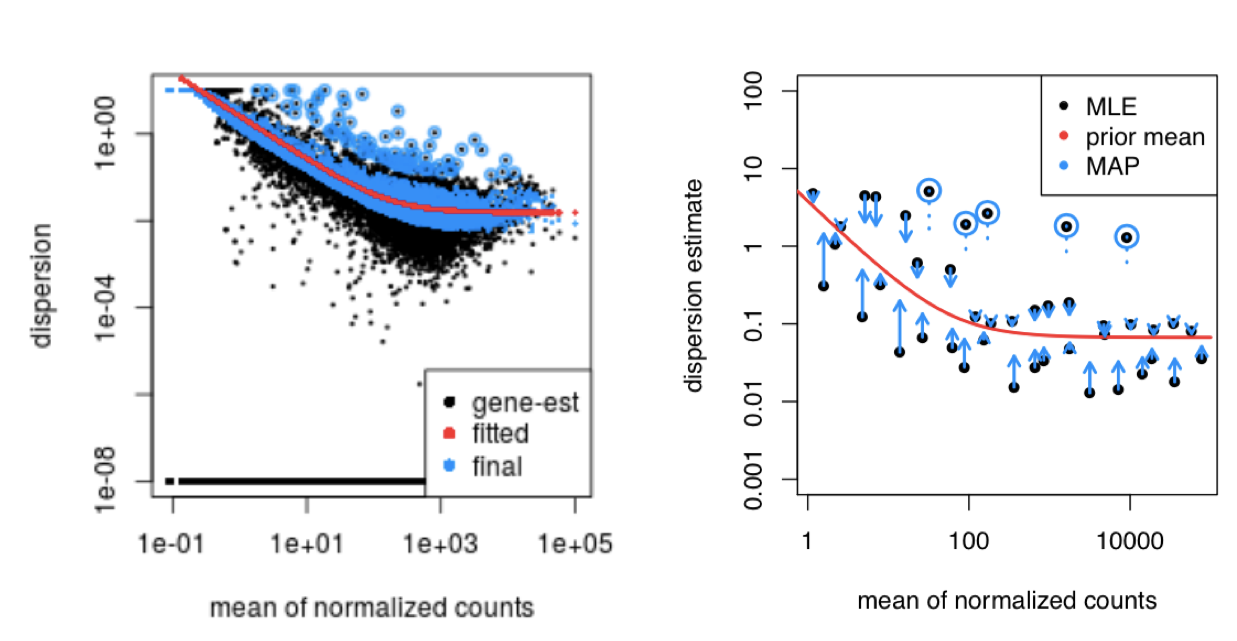
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2 | Genome Biology | Full Text
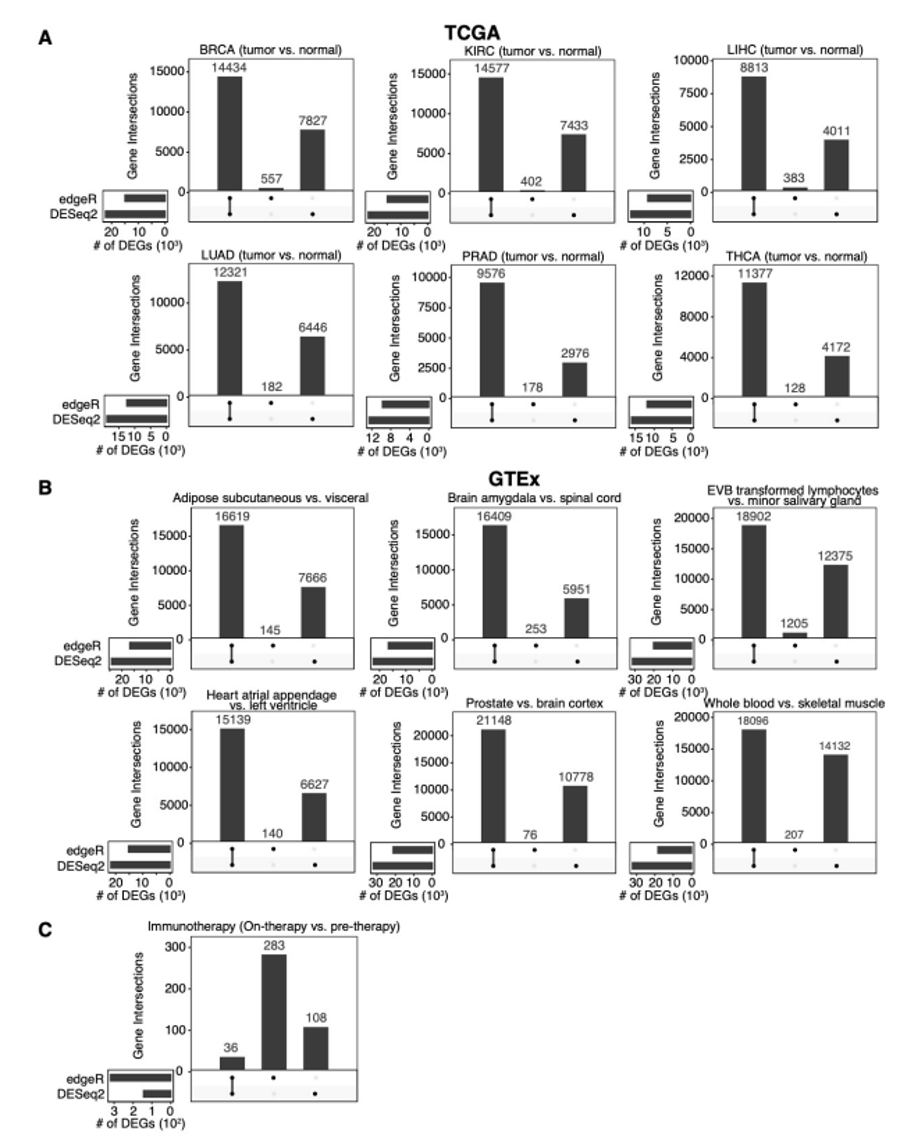
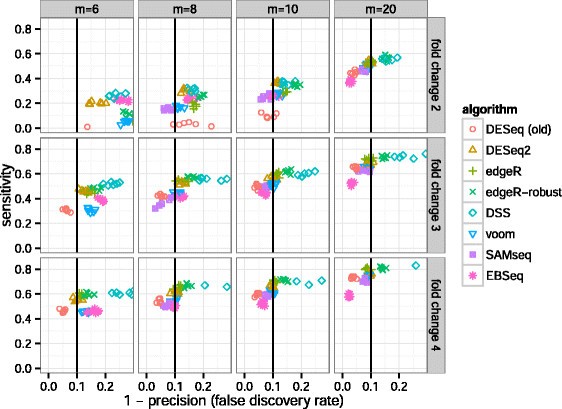
DESeq2 and edgeR should no longer be the default choices for large-sample differential gene expression analysis | by Jingyi Jessica Li | Towards Data Science
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2 | Genome Biology | Full Text

Pairwise comparison (DeSeq2 analysis). Variation in abundance OTUs (p <... | Download Scientific Diagram
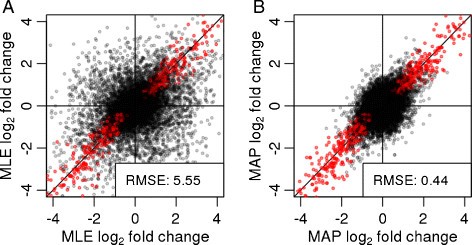
Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2 | Genome Biology | Full Text
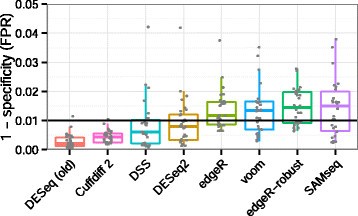
DESeq2 and edgeR should no longer be the default choices for large-sample differential gene expression analysis | by Jingyi Jessica Li | Towards Data Science

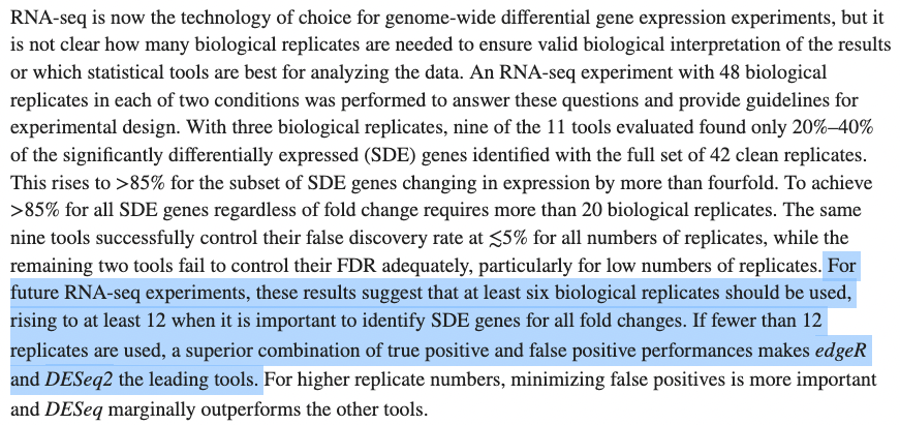
DESeq2 and edgeR should no longer be the default choices for large-sample differential gene expression analysis | by Jingyi Jessica Li | Towards Data Science